

Our Mission
The Microbiome Green Revolution project was developed to tackle a key challenge of the 21st century: how to maintain or increase agricultural productivity while reducing the usage of pesticides and fertilisers, as well as its carbon footprint.
The solution we have proposed is to harness the trillions of microbes in soil, many of which are beneficial to plants. In particular, the project aims to identify and engineer the soil microbial communities that confer protection against disease. We are focusing on a globally important crop, wheat, and the fungal pathogen that causes Take-All disease (Gaeumannomyces tritici, Gt), which causes significant reductions in yield (typically >10-20 per cent) in continuous wheat.
We sincerely thank the Michael Uren Foundation for their generous support in advancing this pioneering research programme. By utilising an enhanced understanding of microbial ecosystems, we aim to foster sustainable agricultural practices and provide farmers with innovative tools to improve crop yields while preserving biodiversity.
Our Research
The research has been designed as an interdisciplinary pipeline that combines state-of-the-art modelling and empirical tools. Figure 1 presents the project’s framework illustrating the connections among the tasks along with staff contributions. Specifically: Task 1 will collect agricultural soils from across the UK to create a biobank of soil microbes; Task 2 will perform high-throughput laboratory experiments to identify protective microbial communities and predict their membership; Task 3 will trial these protective communities in the field. These tasks are integrated using bioinformatic tools and theoretical modelling. Cross-cutting tasks involve development of the computational pipelines and theoretical modelling that will be used to interpret the datasets.
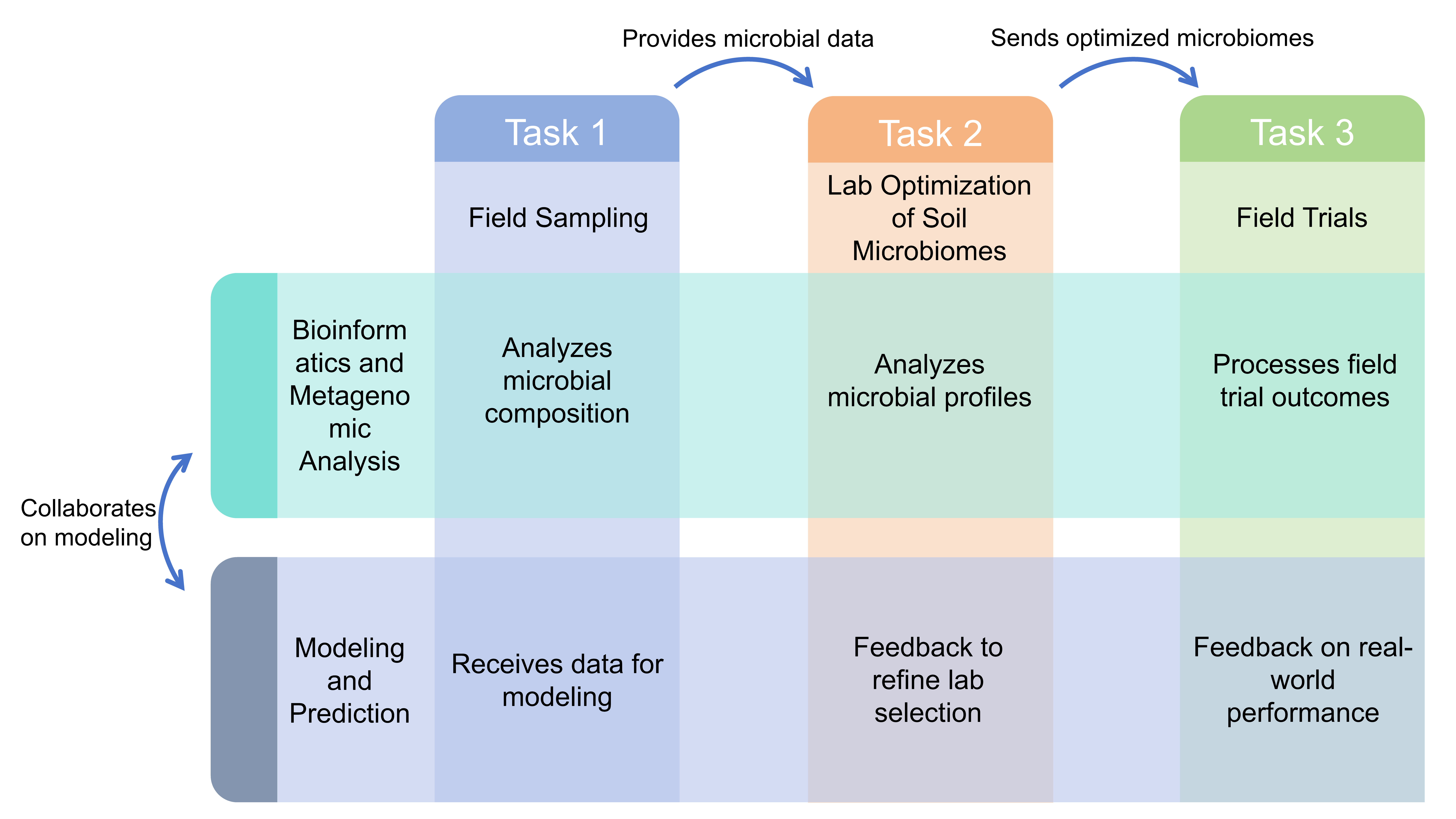
Task 1: Build a biobank from take-all suppressive soils
Our vision is to create a biobank of soils collected from farms across the UK. This biobank would contain the living microbes, kept at cold (4C) or frozen conditions (-80C) to maintain them over the long term. This biobank would be used for the current project and could also be a resource for future projects, for example looking at other plant diseases.
We have finished our first major field campaign, having collected a total of 696 soil samples from across wheat-growing areas of England (see map Figure 2). We have visited farms directly and sent out soil sampling kits for farmers to return by mail. Wheat crops have been harvested over the past few weeks, which has created a natural break in our sampling. We will extend this campaign in the new year.
As part of this work, we have also established collaborations with two leading UK research institutions. Rothamsted Research (rothamsted.ac.uk) is a world-renowned agricultural research institution who will aid with the characterisation of Gt. CABI (cabi.org) is an UN-charter research organisation focused on biocontrol of agricultural pathogens in the developing world. They hold the National Fungal Collection, and a cryo-bank of soils and bacteria associated with UK crops and will assist with characterisation and preservation of the soils
Task 2: Artificial selection of disease-suppressive microbiomes
We will use the Task 1 biobank to conduct a series of experiments that artificially select for the most protective microbial community. Our goal behind using artificial selection is to accelerate the discovery of optimal protocols for self-assembly of beneficial (protective) microbial communities.
We are currently conducting the preliminary experiments required to identify appropriate growing conditions and trial measurement techniques, conducted in our Controlled Environment facility (to be completed in 2024). The main experiments will then begin in Winter/Spring 2025.

Task 3: Test optimised microbiome assembly protocols in the field
The soil microbes and community compositions identified in Tasks 1-2 will be used in a field trial to test their efficacy. This task will be initiated in 2025 following the completion of the first phase of experiments in Task 2.
We have initiated conversations with farmers and are using information collected in the farmer surveys to identify 0best-practice for continuous wheat cropping with minimal risks of Take-all disease.
Development of computational pipelines (cross cutting task)
We are developing robust and reproducible bioinformatic analysis pipelines that will monitor the diverse microbial populations found in the soil samples. Changes in the microbial communities will be compared to wheat yield and resilience to Gt. We are also designing two additional analysis pipelines which will answer:
- Which are the keystone microbes that shape the outcome of the wheat-fungus interaction?
- Which genes help microbes become beneficial symbiotic partners?
- Do viruses play a role in protecting wheat plants from fungal infection?
Future plans development of theory (cross-cutting task)
We are developing the mathematical framework for predicting the incidence of Gt in soils once the datasets are complete. The first component involves statistical modelling of the empirical data collected from the field survey (to be completed by Spring 2025). The second component involves building a ‘digital microbiome’ model of the soil microbiomes (to be completed by end of 2024).